Difference between revisions of "RAW12"
| (50 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
=Format Specifications= | =raw12= | ||
* no header, | ==Format Specifications== | ||
* starting at the top left, with 3072 rows | * no header, little endian, 12bit | ||
* starting at the top left, with 3072 rows of 4096 columns | |||
* the data is in bayer pattern RG/GB | * the data is in bayer pattern RG/GB | ||
* image sensor registers dump (128 x 16bit, big endian) appended | * optional image sensor registers dump (128 x 16bit, big endian) appended at the end (metadata) | ||
==Samples== | |||
http://files.apertus.org/AXIOM-Beta/snapshots/ | |||
==raw12 Image Viewer== | |||
We created a rather simple demo python program to preview raw12 files. Display is not meant to replace a proper raw development workflow but is meant as a quick and easy way to see what a raw12 file contains. | |||
See [[Raw12 viewer]]. | |||
==Quick debayer== | |||
cat test.raw12 | convert \( -size 4096x3072 -depth 12 gray:- \) \( -clone 0 -crop -1-1 \) \( -clone 0 -crop -1+0 \) \( -clone 0 -crop +0-1 \) -sample 2048x1536 \( -clone 2,3 -average \) -delete 2,3 -swap 0,1 +swap -combine test_color.png | |||
==Conversion to DNG== | |||
Convert RAW12 -> DNG with [http://github.com/apertus-open-source-cinema/misc-tools-utilities/tree/master/raw2dng raw2dng]: | |||
raw2dng file.raw12 | |||
Output will be file.DNG; width is assumed 4096, height is auto-detected from file size. For more options, run raw2dng without any arguments. | |||
==Metadata handling== | |||
Show metadata from a raw12 file (without converting it): | |||
raw2dng file.raw12 --dump-regs | |||
or, with [http://github.com/apertus-open-source-cinema/misc-tools-utilities/tree/master/cmv12000-metadata-reader metadatareader]: | |||
cat image.raw12 | dd bs=256 skip=73728 | ./metadatareader | |||
Extract metadata from raw12 image into a separate register file: | |||
cat image.raw12 | dd bs=256 skip=73728 > some.reg | |||
Add metadata from a separate register file to a raw12 image: | |||
cat image.raw12 some.reg > newimage.raw12 | |||
==Understanding the RAW12 file format== | |||
While reading a RAW12 image file such as one given at [[https://files.apertus.org/AXIOM-Beta/snapshots/clipped_wall-BSProt8282-gainx1-offset2047-80ms-01.raw12]], the following points are to be taken into consideration: | |||
* Each pixel data is stored as 12-bit bayer values. This means that RAW12 files contain pixel data from byte 0. For image size of 4096 x 3072, the file size is thus 4096 x 3072 x 12 = 150994944 bits = 18 MB. | |||
* Information is stored in row major order. Hence the first row is RGRGRGRGRG.... and the second row is GBGBGBGB.... and so on. (for RGGB bayer pattern) | |||
* The sensor can mirror the image vertically. This leads to rearrangement of rows (first row becoming the last row and so on). However, this does not change the column order inside the rows. This will lead to RGGB bayer pattern becoming GBRG (every odd row now has GBGBGBGB... and even rows have RGRGRGRG...). This needs to be taken into consideration when dealing with vertically mirrored / vertically flipped images. | |||
A suggested way to extract the bayer channel values is to read the file as binary and treat the cell values 3 bytes at a time. This will encapsulate the values as [RG][RG][RG]... sets for the odd rows and [GB][GB][GB]... for even rows. | |||
For each such set ([RG] or [GB]), the representation is as follows: | |||
[[File:Bayer_bits_odd.png]] | |||
This representation is for odd rows. On even rows, R is swapped by G and G by B bits (for RGGB). | |||
Here, R11 is the MSB of the 12-bit data for Red channel and consecutively, R0 is the LSB. A similar argument goes for G11 and G0. | |||
For getting values of 12 bit channels, the following is a plausible way: | |||
* For red channel: | |||
RedChannel = First8bits<<4 | (Second8bits & 0xF0)>>4; | |||
* For green channel: | |||
GreenChannel = (Second8bits & 0x0F)<<8 | Third8bits; | |||
As always, Red channel is to be swapped with Green Channel and Green Channel with Blue Channel for even rows. (for RGGB) | |||
==Example Python Code to read raw12 data== | |||
Also see [[Raw12 viewer]]. | |||
Here are three examples how to convert the raw data into an RGB image via simple debayering of the RG/GB bayer pattern: http://vserver.13thfloor.at/Stuff/LAM/ | |||
raw12_raw.py simply uses the raw data as 4096x3072 numpy array and retrieves the four nearest RG/GB values to combine them into RGB data which is then written as ASCII PPM image to stdout. | |||
<pre class="code"> | |||
array = np.fromiter(data, np.int) | |||
raw = np.reshape(array, (RAW_HEIGHT, RAW_WIDTH), 'C') | |||
</pre> | |||
raw12_plan.py breaks down the raw data into 2048x1536 | |||
planar arrays for each of the four RG/GB channels: | |||
<pre class="code"> | |||
plan_r = raw[::2, ::2] | |||
plan_g1 = raw[::2, 1::2] | |||
plan_g2 = raw[1::2, ::2] | |||
plan_b = raw[1::2, 1::2] | |||
</pre> | |||
raw12_flat.py finally flattens the planar arrays and | |||
zips them up to present a sequence of RG/GB values: | |||
<pre class="code"> | |||
flat_r = plan_r.flatten() | |||
flat_g1 = plan_g1.flatten() | |||
flat_g2 = plan_g2.flatten() | |||
flat_b = plan_b.flatten() | |||
seq = zip(flat_r, flat_g1, flat_g2, flat_b) | |||
</pre> | |||
Note that you can use each of them on the command line | |||
in the following way ... | |||
<pre class="code"> | |||
./raw12_raw.py some_file.raw12 >some_file.ppm | |||
./raw12_plan.py some_file.raw12 >some_file.ppm | |||
./raw12_flat.py some_file.raw12 >some_file.ppm | |||
</pre> | |||
... and they should give you a PPM image you can open | |||
with your favourite image viewer or image processing | |||
tool. | |||
<pre class="code"> | |||
import itertools | |||
RAW_WIDTH = 4096 | |||
RAW_HEIGHT = 3072 | |||
NUM_REGS = 128 | |||
def raw12_reader(path): | |||
with open(path, 'rb') as f: | |||
for row in range(RAW_HEIGHT): | |||
for col in range(RAW_WIDTH >> 1): | |||
val = f.read(3) | |||
yield (val[0] << 4) | (val[1] >> 4) | |||
yield ((val[1] & 0xF) << 8) | val[2] | |||
for reg in range(NUM_REGS): | |||
val = f.read(2) | |||
yield (val[1] << 8) | val[0] | |||
reader = raw12_reader(raw12_file) | |||
data = it.islice(reader, RAW_WIDTH * RAW_HEIGHT) | |||
regs = it.islice(reader, NUM_REGS) | |||
</pre> | |||
=raw16= | |||
This format is deprecated in favour of the raw12 format above. | |||
==Format Specifications== | |||
* no header, little endian, 16bit (12msb padded with 4lsb zeroes) | |||
* starting at the top left, with 3072 rows of 4096 columns | |||
* the data is in bayer pattern RG/GB | |||
* optional image sensor registers dump (128 x 16bit, big endian) appended (metadata) | |||
RAW12 uses less space than RAW16 but contains identical image data and has no downsides. So we recommend using RAW12 instead (see below). | |||
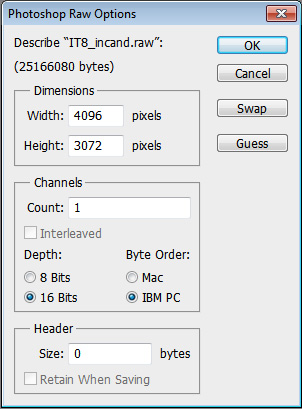
==Opening in Photoshop== | |||
*rename the file extension from ".raw16" to ".raw" | |||
[[File:Raw16-ps.jpg]] | [[File:Raw16-ps.jpg]] | ||
==Samples== | ==Samples== | ||
http://vserver.13thfloor.at/Stuff/AXIOM/RAW/ | |||
XZ compressed sample files (use 7Zip to uncompress under windows: http://www.7-zip.org/): | |||
http://vserver.13thfloor.at/Stuff/AXIOM/ALPHA/RAW/ | |||
==Metadata block== | |||
To display the metadata (optional image sensor registers dump (128 x 16bit, big endian) appended) from a RAW16 file: | |||
cat image.raw16 | dd bs=256 skip=98304 | ./metadatareader | |||
The DNG converter expects the register block to be present so if it has not been saved with the image in camera: | |||
To append an empty metadata block (sensor registers): | |||
(cat old.raw16; dd if=/dev/zero bs=256 count=1) > new.raw16 | |||
To append an existing register block dumped into a file: | |||
cat old.raw16 some.reg > new.raw16 | |||
Latest revision as of 10:53, 30 November 2021
1 raw12
1.1 Format Specifications
- no header, little endian, 12bit
- starting at the top left, with 3072 rows of 4096 columns
- the data is in bayer pattern RG/GB
- optional image sensor registers dump (128 x 16bit, big endian) appended at the end (metadata)
1.2 Samples
http://files.apertus.org/AXIOM-Beta/snapshots/
1.3 raw12 Image Viewer
We created a rather simple demo python program to preview raw12 files. Display is not meant to replace a proper raw development workflow but is meant as a quick and easy way to see what a raw12 file contains.
See Raw12 viewer.
1.4 Quick debayer
cat test.raw12 | convert \( -size 4096x3072 -depth 12 gray:- \) \( -clone 0 -crop -1-1 \) \( -clone 0 -crop -1+0 \) \( -clone 0 -crop +0-1 \) -sample 2048x1536 \( -clone 2,3 -average \) -delete 2,3 -swap 0,1 +swap -combine test_color.png
1.5 Conversion to DNG
Convert RAW12 -> DNG with raw2dng:
raw2dng file.raw12
Output will be file.DNG; width is assumed 4096, height is auto-detected from file size. For more options, run raw2dng without any arguments.
1.6 Metadata handling
Show metadata from a raw12 file (without converting it):
raw2dng file.raw12 --dump-regs
or, with metadatareader:
cat image.raw12 | dd bs=256 skip=73728 | ./metadatareader
Extract metadata from raw12 image into a separate register file:
cat image.raw12 | dd bs=256 skip=73728 > some.reg
Add metadata from a separate register file to a raw12 image:
cat image.raw12 some.reg > newimage.raw12
1.7 Understanding the RAW12 file format
While reading a RAW12 image file such as one given at [[1]], the following points are to be taken into consideration:
- Each pixel data is stored as 12-bit bayer values. This means that RAW12 files contain pixel data from byte 0. For image size of 4096 x 3072, the file size is thus 4096 x 3072 x 12 = 150994944 bits = 18 MB.
- Information is stored in row major order. Hence the first row is RGRGRGRGRG.... and the second row is GBGBGBGB.... and so on. (for RGGB bayer pattern)
- The sensor can mirror the image vertically. This leads to rearrangement of rows (first row becoming the last row and so on). However, this does not change the column order inside the rows. This will lead to RGGB bayer pattern becoming GBRG (every odd row now has GBGBGBGB... and even rows have RGRGRGRG...). This needs to be taken into consideration when dealing with vertically mirrored / vertically flipped images.
A suggested way to extract the bayer channel values is to read the file as binary and treat the cell values 3 bytes at a time. This will encapsulate the values as [RG][RG][RG]... sets for the odd rows and [GB][GB][GB]... for even rows. For each such set ([RG] or [GB]), the representation is as follows:
This representation is for odd rows. On even rows, R is swapped by G and G by B bits (for RGGB). Here, R11 is the MSB of the 12-bit data for Red channel and consecutively, R0 is the LSB. A similar argument goes for G11 and G0.
For getting values of 12 bit channels, the following is a plausible way:
- For red channel:
RedChannel = First8bits<<4 | (Second8bits & 0xF0)>>4;
- For green channel:
GreenChannel = (Second8bits & 0x0F)<<8 | Third8bits;
As always, Red channel is to be swapped with Green Channel and Green Channel with Blue Channel for even rows. (for RGGB)
1.8 Example Python Code to read raw12 data
Also see Raw12 viewer.
Here are three examples how to convert the raw data into an RGB image via simple debayering of the RG/GB bayer pattern: http://vserver.13thfloor.at/Stuff/LAM/
raw12_raw.py simply uses the raw data as 4096x3072 numpy array and retrieves the four nearest RG/GB values to combine them into RGB data which is then written as ASCII PPM image to stdout.
array = np.fromiter(data, np.int)
raw = np.reshape(array, (RAW_HEIGHT, RAW_WIDTH), 'C')
raw12_plan.py breaks down the raw data into 2048x1536 planar arrays for each of the four RG/GB channels:
plan_r = raw[::2, ::2]
plan_g1 = raw[::2, 1::2]
plan_g2 = raw[1::2, ::2]
plan_b = raw[1::2, 1::2]
raw12_flat.py finally flattens the planar arrays and zips them up to present a sequence of RG/GB values:
flat_r = plan_r.flatten()
flat_g1 = plan_g1.flatten()
flat_g2 = plan_g2.flatten()
flat_b = plan_b.flatten()
seq = zip(flat_r, flat_g1, flat_g2, flat_b)
Note that you can use each of them on the command line in the following way ...
./raw12_raw.py some_file.raw12 >some_file.ppm
./raw12_plan.py some_file.raw12 >some_file.ppm
./raw12_flat.py some_file.raw12 >some_file.ppm
... and they should give you a PPM image you can open with your favourite image viewer or image processing tool.
import itertools
RAW_WIDTH = 4096
RAW_HEIGHT = 3072
NUM_REGS = 128
def raw12_reader(path):
with open(path, 'rb') as f:
for row in range(RAW_HEIGHT):
for col in range(RAW_WIDTH >> 1):
val = f.read(3)
yield (val[0] << 4) | (val[1] >> 4)
yield ((val[1] & 0xF) << 8) | val[2]
for reg in range(NUM_REGS):
val = f.read(2)
yield (val[1] << 8) | val[0]
reader = raw12_reader(raw12_file)
data = it.islice(reader, RAW_WIDTH * RAW_HEIGHT)
regs = it.islice(reader, NUM_REGS)
2 raw16
This format is deprecated in favour of the raw12 format above.
2.1 Format Specifications
- no header, little endian, 16bit (12msb padded with 4lsb zeroes)
- starting at the top left, with 3072 rows of 4096 columns
- the data is in bayer pattern RG/GB
- optional image sensor registers dump (128 x 16bit, big endian) appended (metadata)
RAW12 uses less space than RAW16 but contains identical image data and has no downsides. So we recommend using RAW12 instead (see below).
2.2 Opening in Photoshop
- rename the file extension from ".raw16" to ".raw"
2.3 Samples
XZ compressed sample files (use 7Zip to uncompress under windows: http://www.7-zip.org/):
http://vserver.13thfloor.at/Stuff/AXIOM/ALPHA/RAW/
2.4 Metadata block
To display the metadata (optional image sensor registers dump (128 x 16bit, big endian) appended) from a RAW16 file:
cat image.raw16 | dd bs=256 skip=98304 | ./metadatareader
The DNG converter expects the register block to be present so if it has not been saved with the image in camera:
To append an empty metadata block (sensor registers):
(cat old.raw16; dd if=/dev/zero bs=256 count=1) > new.raw16
To append an existing register block dumped into a file:
cat old.raw16 some.reg > new.raw16